‹ Go back to the installation page
DeepInfer usage guide
DeepInfer can be used in command-line mode or through 3D Slicer. In the following sections we describe both methods:
DeepInfer 3D-Slicer
Make sure that you have installed both 3D Slicer and DeepInfer module and have validated your installation. Follow the steps bellow to connect a model from DeepInfer model store and run it on your machine:
-
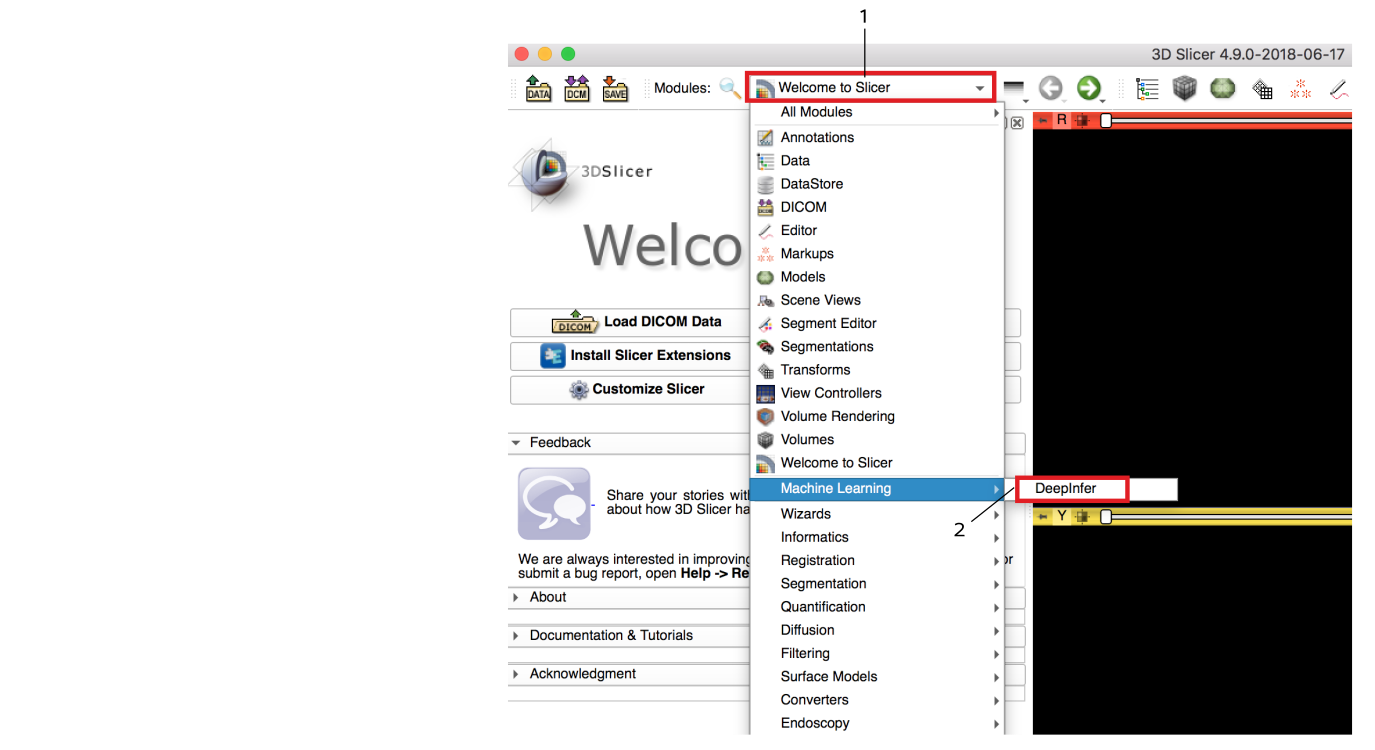
Open DeepInfer module: Go to the modules menu and choose Machine Learning > DeepInfer module.
-
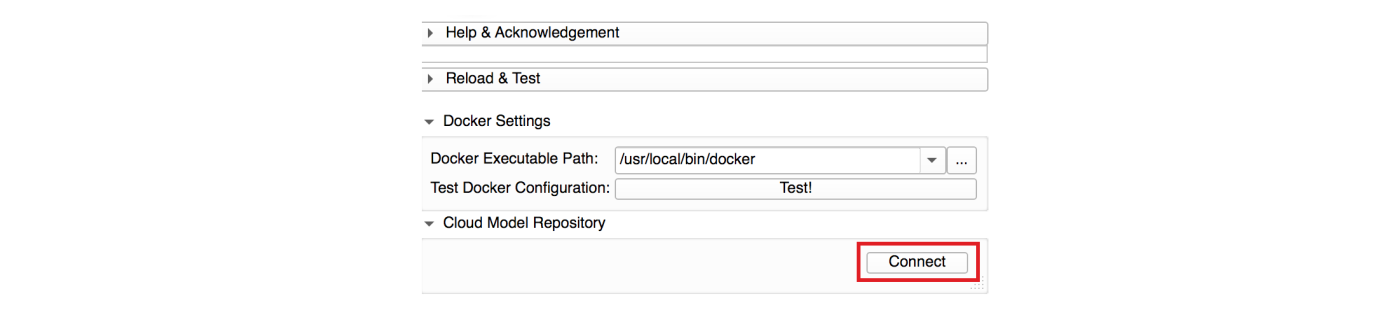
Connect to the DeepInfer Model Store: make sure that your computer is connected to the internet. Press the connect button under the Cloud Model Repository tab. It will connect to DeepInfer model store and creates a list of available models.
-
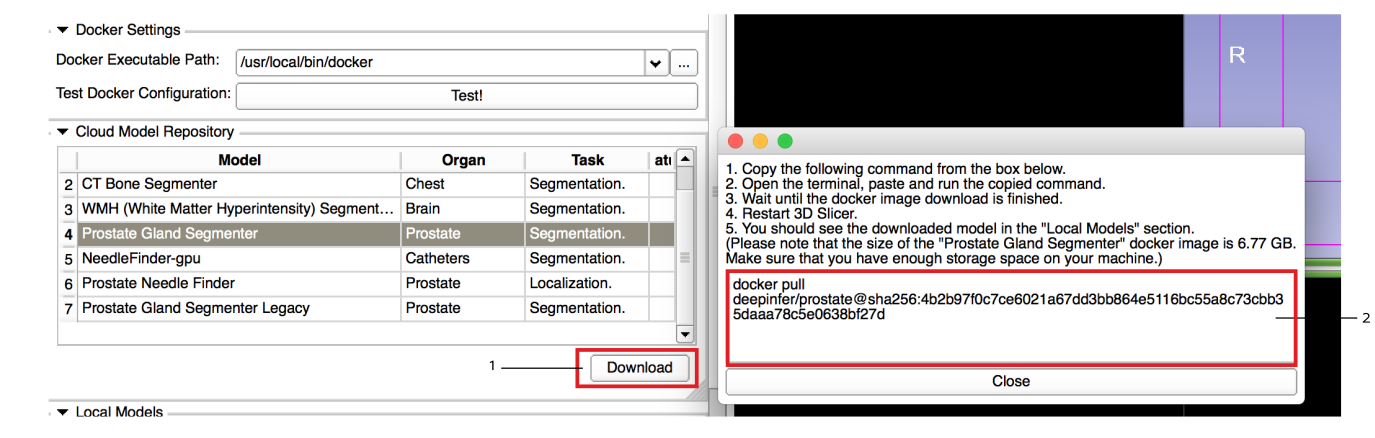
Select and download your selected model(s): from the populated table select a model and press the download button. This will prompt a popup window with a terminal command to download (pull) the docker image. Copy and paste the command in a terminal and wait until the download is finished. Once the download is finished, restart Slicer and you should see the model in the local models section.
-
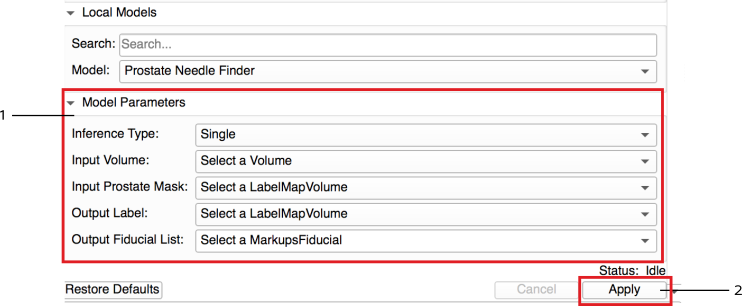
Set input(s) and output(s) and run the inference: Load the input images and set the input nodes with appropriate inputs. For output nodes make sure to create new nodes. Once you create all nodes press the "Apply" button. A progress bar will appear wait until the process is finished and progress reaches to 100%.
Resetting the module If for any reason the inference procedure breaks, reset the module to its initial condition by pressing the reload button under "Reload & Test".
DeepInfer Command-line
DeepInfer models can be run outside of 3D slicer in the command-line mode. To run the model in
command-line,
check the specific documentation page of the model from the model store for instructions.
Here is an example of downloading and running the prostate-segmenter model in command line mode:
-
Download the docker image
docker pull deepinfer/deepinfer-prostate -
Run the docker: By running the following code we first map a folder in the file system to the data folder inside the docker, and the giving the input volume name, output prediction label path and other model parameters. After running the following command the predicted result will be generated inside the mapped folder.
docker run -t -v ~/data/prostate_test/:/data deepinfer/prostate\ --ModelName prostate-segmenter\ --Domain BWH_WITHOUT_ERC\ --InputVolume /data/prostate.nrrd \ --OutputLabel /data/output_prostate_label.nrrd \ --ProcessingType Fast\ --Inference Ensemble\ --verbose
For other models check the "Command-line interface guide section" in their model page.